How to Submit R Jobs on Hoffman2 Cluster¶
Pre-requisites: a Hoffman2 account.
Helpful readings: Googling basic shell commands for naviaging the file system, e.g. ls (list files), cd SubDirectoryNAME (go to a subfolder, back to home, or go back to parent folder cd ..), cp (copy), etc.
After logging in, you should be in your home directory, with no files at all or one or two files. Now let's create a directory (folder) called "testDir".
(I'm demonstrating BASH Shell code in Jupyter Notebook, which requires the ! in the front. In practice, ignore the !. Just type in the command, and press ENTER)
#Go Home
!cd
#Create a directory called "test"
!mkdir testDir
#List all files here. You should be able to see your new folder, "test"
!ls
Copying your R script to cluster¶
I am using @hoffman2.dtn.hoffman2.idre.ucla.edu domain, per UCLA Hoffman2 policies for large file transfers. You can use @hoffman2.idre.ucla.edu if your file isn't large, such as our case, an R script. You can do this in a separate terminal, without logging out of Hoffman2.
#Don't forget to first navigate, in a terminal, to the folder that contains your script on your machine first!
!scp YOUR_R_SCRIPT YOUR_USERNAME@hoffman2.dtn.hoffman2.idre.ucla.edu:testDir/
#Or this, if you just want to put the file in your home directorym instead of a folder
!scp YOUR_R_SCRIPT YOUR_USERNAME@hoffman2.dtn.hoffman2.idre.ucla.edu:.
Now go to your Hoffman2 folder testDir, you should see your new R script
!ls testDir/.
# this is equivalent to, cd testDir ENTER and ls
Install Required R Packages for Your Scripts.¶
You have to do the following steps to set up required packages for your R script. Good news is that you only have to do this once.
Inside my test.R file, I only wrote two lines of code library(dplyr), and print("Runing my first Hoffman2 job.") (You may do the same if you are practicing along with this tutorial). However, before submitting the job, we need to make sure dplyr package is installed. In the future, you need to do this (just once for each new package) for all package you will be using in the script.
BTW, You may edit test.R directly in terminal using vi or nano, etc. editors. Please Google these documentations to learn how. Otherwise, you can either scp upload each time, as described above, or use Jupyter Notebook UI to edit directly on the cluster (See my future tutorials).
#Load Correct R version from Hoffman2. For example,
module load R/4.0.2
# If you are not sure what versions that Hoffman2 has, check all available softwares:
module avail
#Launch R in terminal
!R
#Once inside R, write
install.packages("dplyr")
IMPORTANT: The R program will tell you that you don't have root access, asking you if you want to install it in your local directory. Remember to save/copy-paste this path. Type in y (yes), and enter. If you forget this path later, don't worry. Just launch R again, and run .libPaths(). This will show you two paths. The second one is your personal R library directory. This path should look something like, /u/home/SOME_LETTER/YOUR_USERNAME/R/x86_64-pc-linux-gnu-library/4.0.2/.
IMPORTANT: all the R packages you installed above, or will install in the future, correspond to the R version (as you can see from the library path above). This means if you load a different R version module next time you will have a different (empty if you never installed) library. Thus, every time you install, make sure you have loaded the correct R version!
Once you are done, put .libPaths("YOUR SAVED LIB PATH") in front of your every R script, so when Hoffman2 runs your code, it knows where to find the packages that you installed!
Last Step before Submitting, Building a Command File For Submission¶
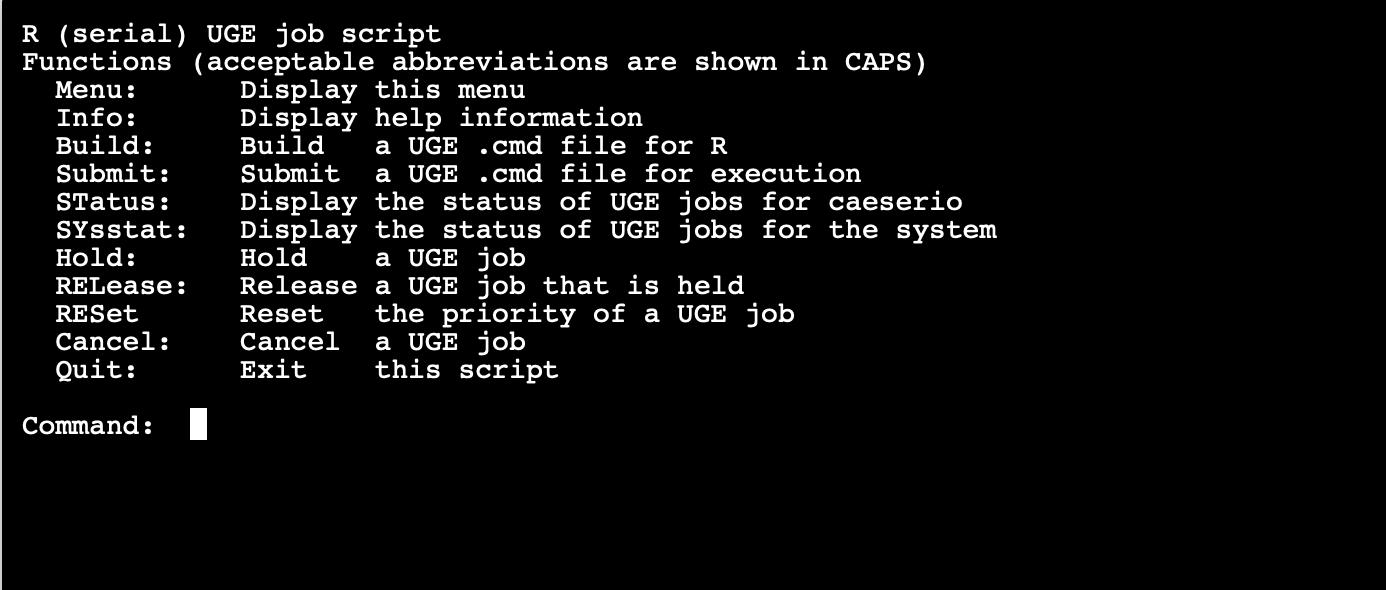
This step seems odd, but it is required for cluster's job submission engine (Hoffman2 uses Sun Grid Engine, or SGE) to work. Normally, you would have to write your own grid engine command file. Luckily, Hoffman2 has a small program written for R jobs. You just need to launch this program R.q (I recommend first navigeting to the folder where the R script is saved, so when you tell the program to build upon R script, you won't have to type in the whole path).
Then the program starts. Just press enter, and type in build.
Then it will ask you a series of questions, the first of which is the name of your R script. If you followed the above instructions and already nagvigated to the folder before launching R.q, just type in the R script name exactly as it is (in our case test.R).
As for questions like how much RAM etc., it depends on how much RAM your code needs. For additional commands you can just leave blank and enter. "Would you like to submit now?" type in "n" (No).
Once you are done, go to this newly created file, which should have exactly the same name as your R script except the extension is ".cmd" in the same directory(folder). Open it via vi or other editors. Find this part where it says, source /u/local/Modules/default/init/modules.csh
module load R (image below). Replace module load R with module load R/YOUR_Version, for example in our case, module load R/4.0.2. This way, when the cluster runs your job, it will load the correct R version.

Submitting the job¶
Submitting is easy (Reminder to check your R script has .libPaths("YOUR SAVED LIB PATH") in the front). In terminal, simply navigate to where your R script and cmd file are saved, and type in qsub test.cmd
!qsub testDir/test.cmd